3
The story behind our new paper in PeerJ
Oct 14, 2013
1632 views
(Reposted with permission from Todd Vision's laboratory website)
The paper contributes to the evidence base about the costs and benefits to researchers for archiving their data publicly at the time of publication. We focused on one of the key potential benefits, increasing citations to your paper. There have been a variety of correlational studies that find a citation benefit for articles when the underlying data is publicly archived, including one from first-author Heather Piwowar in 2007 which found a very large effect (with correspondingly large error bars, since it looked at only 85 articles). But since these sorts of studies do not randomly allocate researchers to open/non-open data treatments, there is always the concern that the measured benefit could be driven by confounding variables. For instance, if the study was conducted by a single investigator versus a consortium, this could affect both whether the data is publicly archived and how many citations it receives.
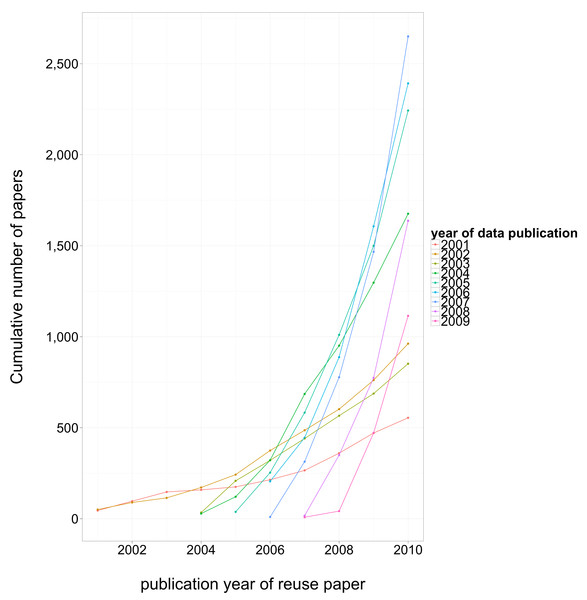
Our new study, while still correlational, has a sufficiently large sample (over ten thousand publications), that multivariate analysis can effectively factor out a wide range of potential confounding variables. Like Heather’s 2007 study, the papers we looked at are all reporting gene expression data, and a comparison is possible because only 25% of them openly archived their data in one of the major gene expression repositories (GEO and ArrayExpress). We found, after accounting for other factors, on average a 9% citation benefit from open archiving, with a higher benefit (up to 30%) for earlier years (where more citations overall had accumulated). Manual inspection of a small subset of the citing literature suggests that much of the benefit appears to be driven by other researchers actually reusing the data.
One finding of relevance to data archiving policy was how long it took for authors to cease publishing papers reusing their own previously published data. We found that reuse dropped off steeply and was negligible after two years. This reinforces the hunch, enshrined in policies such as JDAP, that embargoing data for more than about a year post-publication may do more to harm the citation benefit to the original authors more than to help protect any proprietary interest they have in the data. Though, it would still be nice to have evidence from this for other datatypes in biology, and indeed from other disciplines.
The fascinating but maddening story behind Heather’s attempts to get text-mining access to the literature in order to detect these trends is alluded to in the Discussion. For more of the story, worth a blog post its own right, see this excellent piece in the Guardian.
Publishing this paper with PeerJ has been a fun ride, both because the staff have been so responsive and the platform is so well-designed, but also because we have had a chance to test-drive new features as they are introduced. Our unreviewed manuscript was the first PeerJ PrePrint, and now that it has been published, we have already started interacting with readers via PeerJ’s innovative new Q&A; feature.
But wait, there’s more! The data are available in Dryad for reuse by others. In fact, the executable manuscript is available there, too. The whole thing was written using Knitr on GitHub with embedded R code and data, so the analyses in the paper, including tables and figures, can be updated just by modifying and replacing the data and recompiling the thing. Furthermore, the peer reviews (in one case, even signed) and our response letter have been made openly available alongside the PeerJ article.
At the time of this study, Heather was a postdoc supported by DataONE. She is now a postdoc emeritus working full-time on her nonprofit altmetrics startup ImpactStory. There are still a couple other exciting papers on the way from her DataONE work, so stay tuned.
Finally, since Heather and I both are more than a little interested in altmetrics, it’s fun to watch how much viewership the article has gotten so far, over 1,000 unique visitors in less than a week, and how much of the traffic has been driven by stories in the popular science press. That includes a story in the online edition of Spiegel, which quotes me saying things in German that are far beyond my abilities in that formidable tongue.
Copyright: © 2013 Todd Vision. The above content is licensed under the Creative Commons Attribution License (CC-BY), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. 

If you find this essay offensive or in violation of your rights, please email to [email protected]

COMMENTS